Whole genome duplication or polyploidization produces organisms (i.e., polyploids) with three or more sets of chromosomes, a process that has been the hallmark of plant evolution and shaped the genomes of all living plants, including most of our crops. It continues to be responsible for a sizeable percentage of new species on the planet today. A group of researchers, led by Ovidiu Paun from the Department of Botany and Biodiversity Research of the University of Vienna, and including researchers from the UK, Czech Republic, Sweden and Australia, adds an important missing piece to our understanding of genome evolution after polyploidization. They investigated five wild polyploid marsh orchids of increasing ages, ranging from ca 500 to ca 100,000 generations.
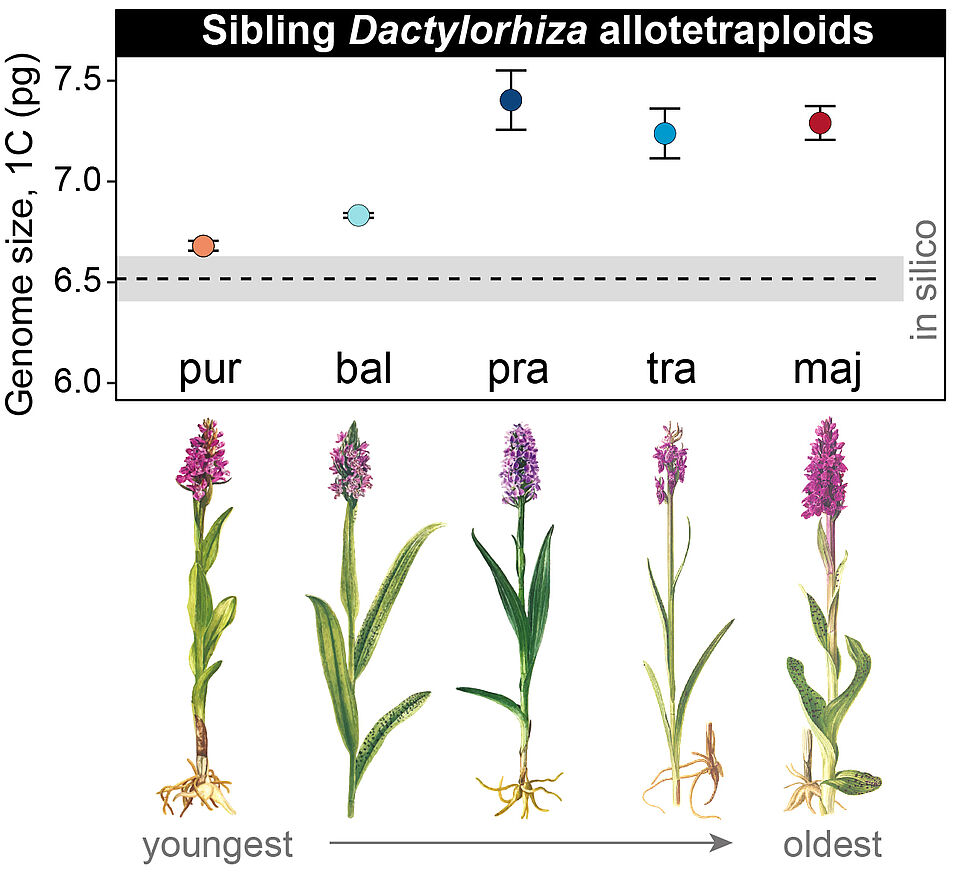
Previous studies, mainly done on laboratory strains, established that polyploidy often activates dormant selfish genomic components (called transposable elements) that can jump or multiply within the genome and trigger genome size increases and restructuring. The current study, funded by a START grant of the Austrian Science Fund (FWF) used wild species of different ages to view “evolution in time”, in contrast to using artificially produced organisms. They investigated five sibling marsh orchids, who have a relatively large genome size, about 55 times larger than the model plant Arabidopsis. The different siblings derive from the same two parent species, the common spotted orchid, Dactylorhiza fuchsii, and the early marsh orchid, Dactylorhiza incarnata and formed sequentially as a result of similar crossings. The study made use of the increasing ages of the five polyploids to generate a perspective through time of the genome dynamics post-polyploidization, which allowed to test the theory that an increase in transposable element activity should quickly follow polyploid formation.
Five sibling Orchids provide a window into genomic evolution
The researchers found that the youngest sibling, the northern marsh orchid, Dactylorhiza purpurella (endemic to the British Isles and Denmark), has a genome size and repeat content close to the sum of the parental genomes. In contrast, polyploids of intermediate ages, the eastern European Dactylorhiza baltica and western European Dactylorhiza praetermissa (southern marsh orchid) exhibited a sharp increase in genome size. The oldest polyploids in the series, including the narrow-leaved marsh orchid Dactylorhiza traunsteineri and the broad-leaved marsh orchid Dactylorhiza majalis, are similar in size to those of the intermediate ages. ” This indicates that activation of transposable elements does not immediately follow polyploidization and stabilizes relatively quickly after a period of rapid increase”, says Ovidiu Paun, the lead author of the study.
'Genomic Shock' does not cause multiplication events
The observed increase in genome size in the polyploid marsh orchids happens on a magnitude of multiple entire genomes of some plants such as the model-plant Arabidopsis. Most of this increase is specifically driven by one particular transposable element, meaning that the great majority of repeats remain strongly regulated and therefore their fate is predictable across polyploidization events in this system. This speaks against the hypothesis of a generalized genomic shock triggering widespread multiplication of repetitive elements, a concept put forward by Barbara McClintock in 1984. The absence of a major, global activation of transposable elements in polyploid marsh orchids may be linked to their large genome sizes, which could expose them to strong selection against even further genome expansion.
Publication in Molecular Biology and Evolution:
Eriksson MC, Mandakova T, McCann J, Temsch EM, Chase MW, Hedren M, Weiss-Schneeweiss H, Paun O. 2022. Repeat dynamics across timescales: a perspective from sibling allotetraploid marsh orchids (Dactylorhiza majalis s.l). Molecular Biology and Evolution in press.
https://doi.org/10.1093/molbev/msac167
Scientific contact:
Ovidiu Paun
Botany and Biodiversity Research
Faculty of Life Sciences, University of Vienna
Rennweg 14, 1030 Vienna.
Tel +436763126637
ovidiu.paun@univie.ac.at
https://plantgenomics.univie.ac.at